Reaction path calculations allow the study of the intricate details of complexation mechanisms. One can actually observe the dynamics of complexation.
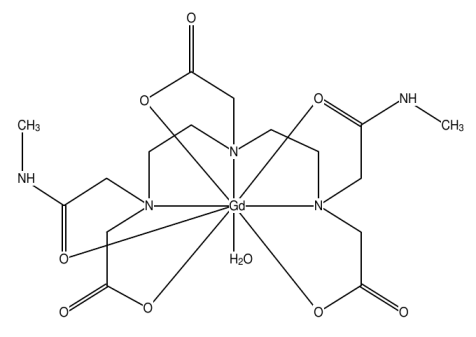
In this tutorial, we will compute the reaction path for the coordination of the water molecule in UDOMIJ [Aqua-(diethylenetriamine-bis(acetic acid methylamide)triacetato)-gadolinium] below:
- Draw the complex in GABEDIT as described in the Drawing Complexes tutorial up to the pre-optimization of the geometry with molecular mechanics. Do not optimize the geometry with MOPAC2012 as of yet. We need to perform a simple procedure before that, as described below:
- Right-click anywhere on the window "Gabedit: Draw Geometry", and then choose "Edit" > "Open XYZ or GZMAT editor". Find the lanthanide atom in the complex and set its number in column "Nr" to 1. This will place the lanthanide atom in the first position of the internal coordinates of the complex.
- Now, you may optimize the geometry as described in Optimize the geometry of the lanthanide complex with MOPAC2012.
- Right-click anywhere on the window "Gabedit: Draw Geometry", and then choose "Edit" > "Open XYZ or GZMAT editor". Find the lanthanide atom in the complex and set its number in column "Nr" to 1. This will place the lanthanide atom in the first position of the internal coordinates of the complex.
- Go to the directory where you saved the files. Open the .arc file and save it with another name with the .mop extension. Find the line which reads FINAL GEOMETRY OBTAINED and remove everything above and including this line.
- Replace the keywords present by the followng keywords AM1 INT 0SCF SPARKLE CHARGE=0 Singlet. We set CHARGE=0 because that is the charge of UDOMIJ. For other complexes, please set the CHARGE keyword to the appropriate value.
- Save the file as a .mop file with another name. For convenience we provide udomij_opt.mop which contains the optimized geometry of UDOMIJ
- Run this new .mop file in MOPAC2012. The effect of this calculation will be to change the geometry of the molecule from cartesian to internal cordinates. In this case, we need internal coordinates to be able to run reaction path calculations for complexes of lanthanides.
- Open the .arc file which resulted from this last calculation and save it with another name with the .mop extension. Find the line which reads FINAL GEOMETRY OBTAINED and remove everything above and including this line.
- Save this file as a .mop file with another name. For convenience we provide udomij_opt_int.mop, which contains the optimized geometry of UDOMIJ in internal coordinates.
- Open the "udomij_opt_int.mop". Each line refers to each atom of the complex. Present in the second column of the file, are the interatomic distances between the atom of that line and the atom whose number is appears in the fifth column of that line.
- In this file, now identify the atoms of the complex diretly coordinated to the lanthanide ion. They will be represented by lines with the number 1 in the fifith column. For them, the interatomic distance appearing in column 2 is the exact distance between this atom and the lanthanide ion.
- Let us now choose the coordinaing atom of a ligand whose complexation reaction pah we want o calculate. For this tutorial we will choose to study the coordination of the water molecule in UDOMIJ. The oxygen directly coordinating to the Gd(III) ion is the 7th atom in line 10 of file udomij_opt_int.mop. The two hydrogen atoms bonded to this oxygen are the 11th and 12th in lines 14 and 15 of file udomij_opt_int.mop.
- Edit the "udomij_opt_int.mop" replacing the number on the third column of line 10 corresponding to the chosen oxygen atom, from +1 to -1. This means that the reaction path will be computed by varying the interatomic distance between the oxygen atom and the lanthanide ion only. The fact that internal coordinates are being used means that the two hydrogen atoms will follow the oxygen during the calculation.
- Replace the keywords, of the .mop file, by the keywords AM1 AUX SPARKLE 1SCF STEP=0.01 POINT=400 GEO-OK CHARGE=0 Singlet. That means that 400 frames will be produced at steps of 0.01Å, as z matrices in the .out file, as well as in another automatically generated file of extension .xyz.
- For convenience, we provide a zipped udomij_opt_int.xyz.zip.
- To visualize a movie of the complexation reaction path, open Gabedit and click on “Display Geometry/Orbitals/Density/Vibration” button:
 . This will open a new window called “Gabedit: Orbitals/Density/Vibration”.
. This will open a new window called “Gabedit: Orbitals/Density/Vibration”.
- Right click on the black screen and choose the option “Animation” > "several geometries (Convergence/IRC)" in the menu that appears. A new window, called "Multiple Geometries", will appear. Go to the "File" option in this window, and select “Read a xyz file” from the drop down menu that apears. In the new window that appears, named "Read geometries form a XYZ file", navigate to the proper .xyz file and open it.
- Now, position the complex as you wish, for an easy view. Notice that a new option will appear in the bottom of the "Multiple Geometries" window called "Play". Click on it to watch the movie. Below, we provide a YouTube version of this movie:
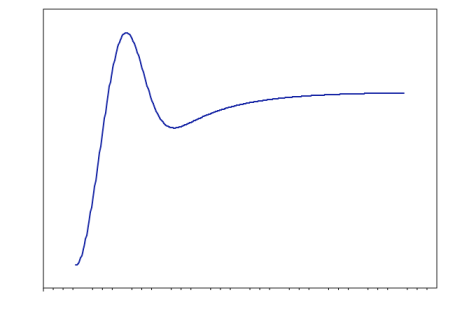
- Now, you may also visualize the Reaction Path Profile of the coordinating water molecules. Please, note that when you run a reaction path Mopac calculation a .arc file will be automatically generated containing the reaction path profile. For convenience, we provide udomij_opt_int.arc.
- Open Gabedit and click on “Tools” > "XY plotter". This will open a new window called “XY GabeditPlot”. Right click on the white screen and choose the option “Data” > "Add Data" > "Read data from an ASCII XY file(2 columns)" in the menu that appears. In the window that appears, make sure you select the option that displays all file types, so that the .arc files appear. Select the appropriate .arc file and click open. The reaction path profile is then displayed, as below: